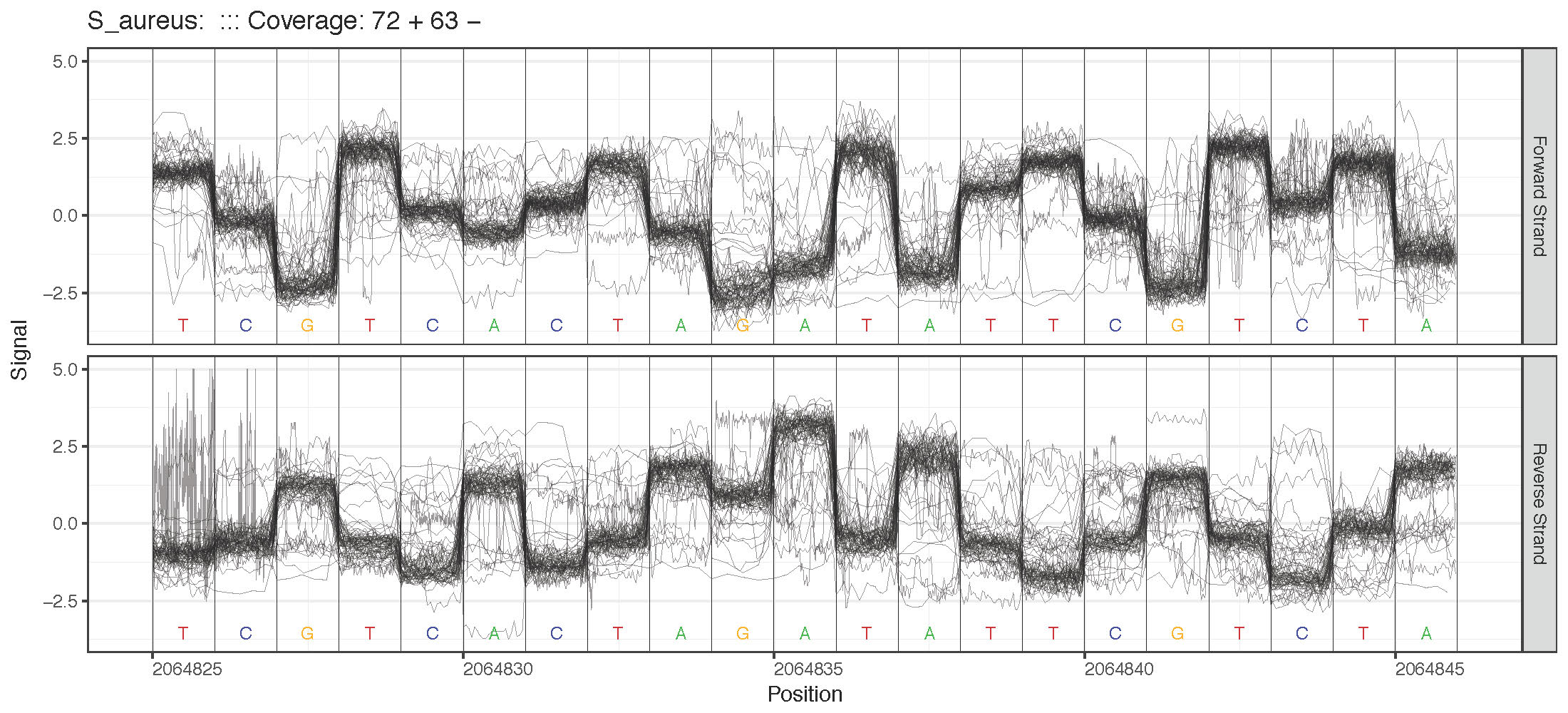
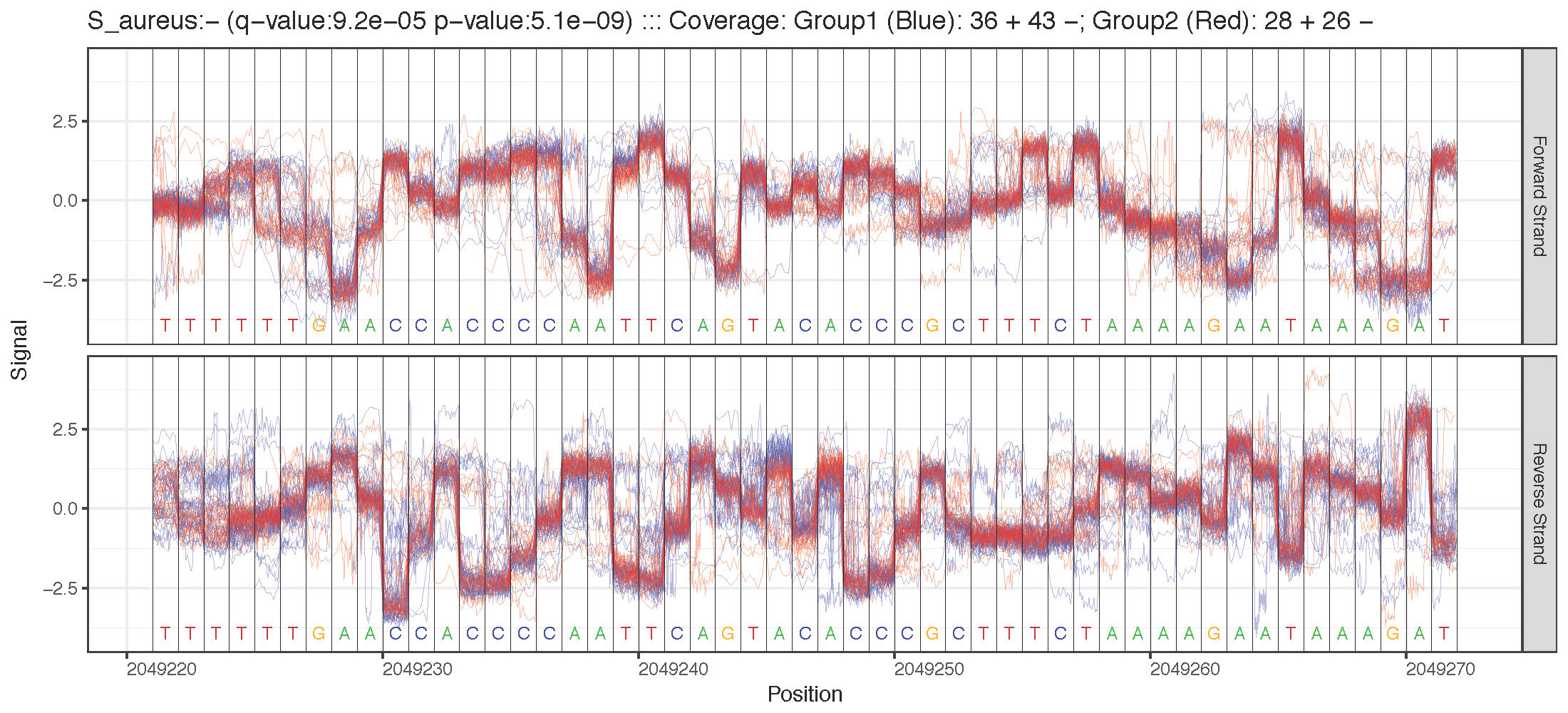
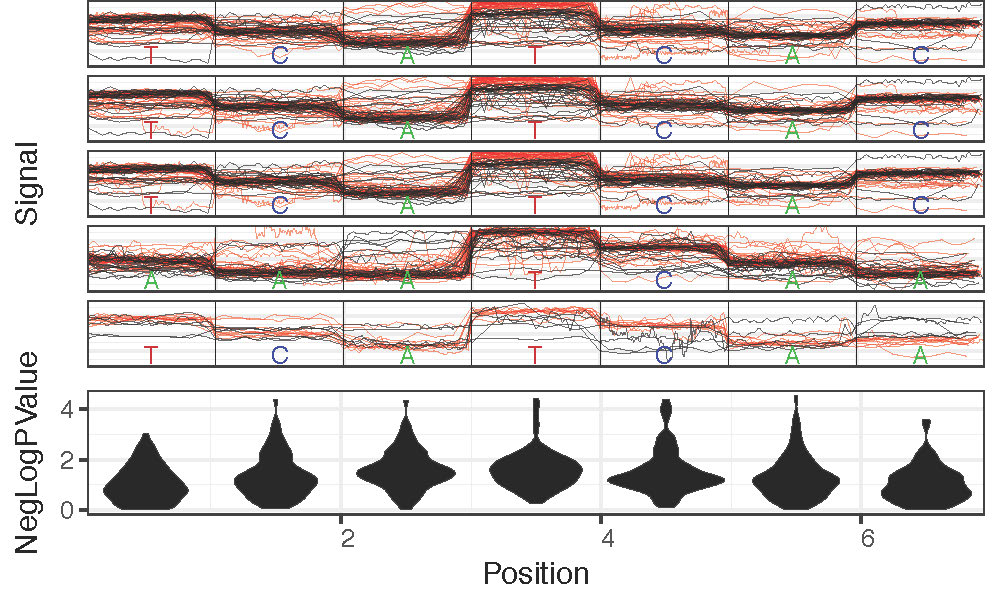
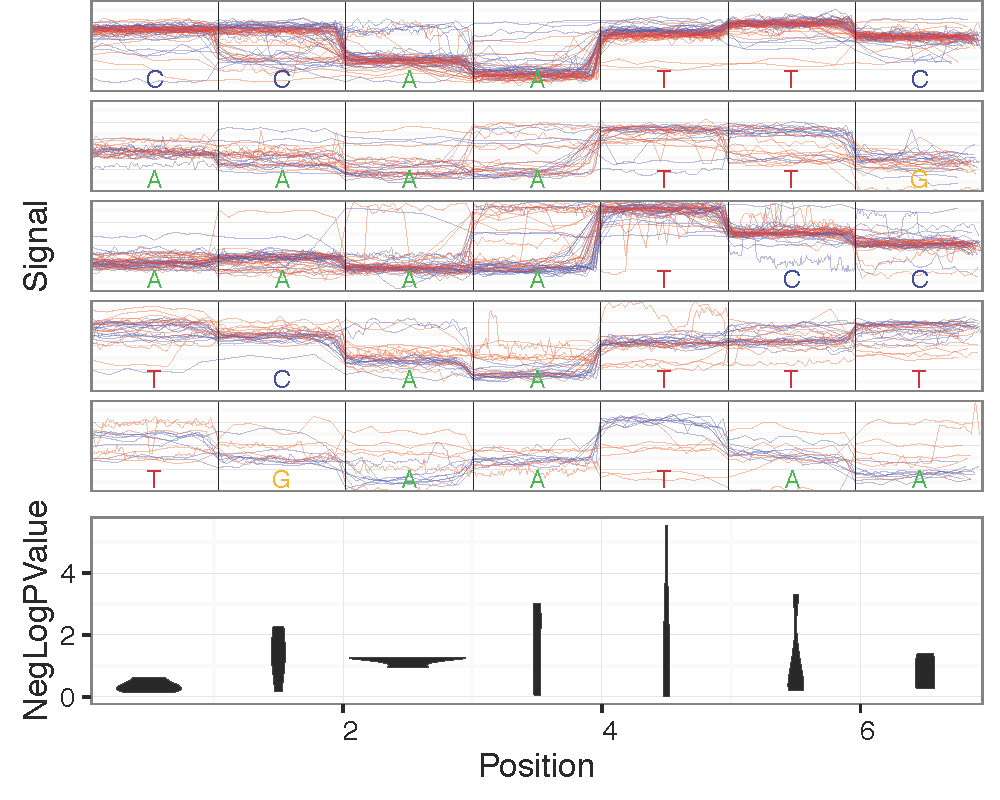
Genome Anchored Plotting Commands¶
Single Sample Plotting¶
Region Plotting Selection Criterion¶
- Maximum coverage (plot_max_coverage)
- Specified Genome Locations (plot_genome_location)
- Centered on motif of interest (plot_motif_centered)
Two Sample Plotting¶
Region Plotting Selection Criterion¶
All those available from single sample plotting plus:
- Maximum difference in mean signal level between two samples (plot_max_difference)
- Most significant statistical test between two samples (plot_most_significant)
Motif Centered Multi-location with Statistics Plotting¶
Plot statistics distrbution around motif of interest across many genomic locations along with most significant example genomic regions with plot_motif_sith_stats.
Common Options¶
- –fast5-basedirs: One or more directories containing FAST5 files that have been “re-squiggled” (nanoraw genome_resquiggle). This option is required for all genomic plotting commands.
- –fast5-basedirs2: One or more directories containing FAST5 files that have been “re-squiggled” (nanoraw genome_resquiggle). These will be group2. This option is required only for plot_max_difference, plot_most_significant and plot_motif_with_stats
- –pdf-filename: Filename to store plots from this command. (Default depends on the command)
- –num-regions: Number of difference regions to plot. Each region will be on another page of the output PDF and ordered by criterion (if applicable). This option is not valid for plot_genome_location.
- –num-bases: Number of genomic bases to include in a plot. Selection criterion will apply to the central base of a plotted region.
- –obs-per-base-filter: Filter reads for plotting baseed on threshold of percentiles (over all bases in a read) of the number of observations assigned to a base. Format thresholds as “percentile:thresh [pctl2:thresh2 …]” E.g. reads with 99th pctl <200 obs and max <5k obs would be “99:200 100:5000”. Default is no filter.
Overplotting Options¶
- –overplot-threshold: Number of reads to trigger alternative plot type instead of raw signal due to overplotting. Default depends on command.
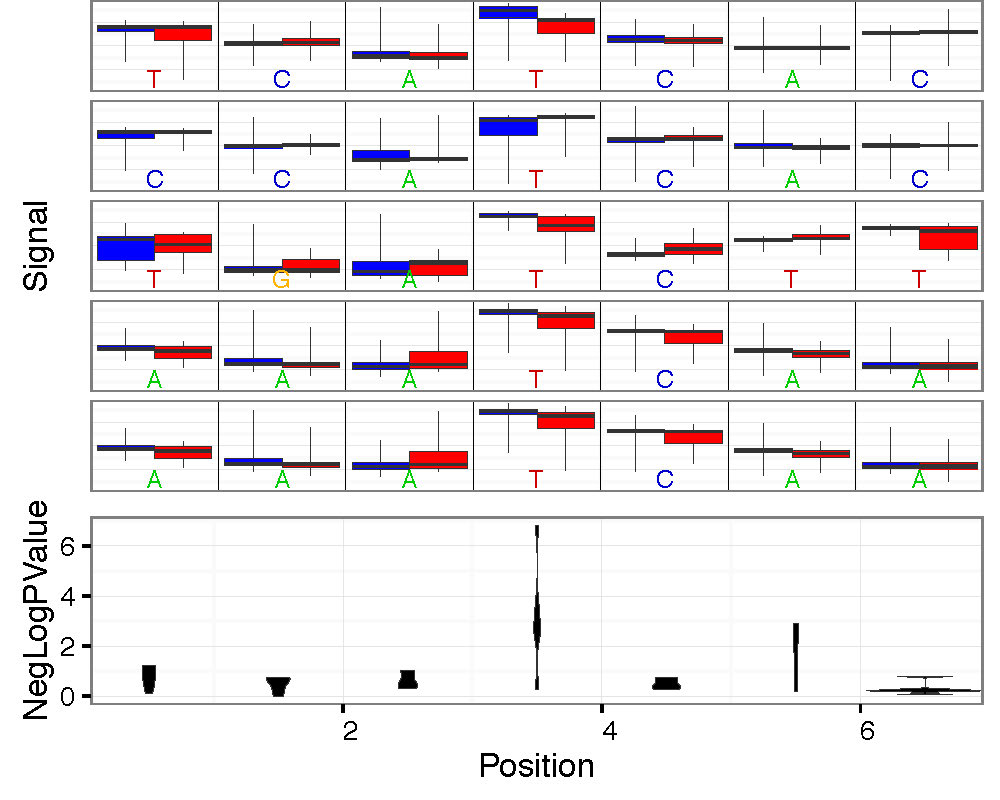
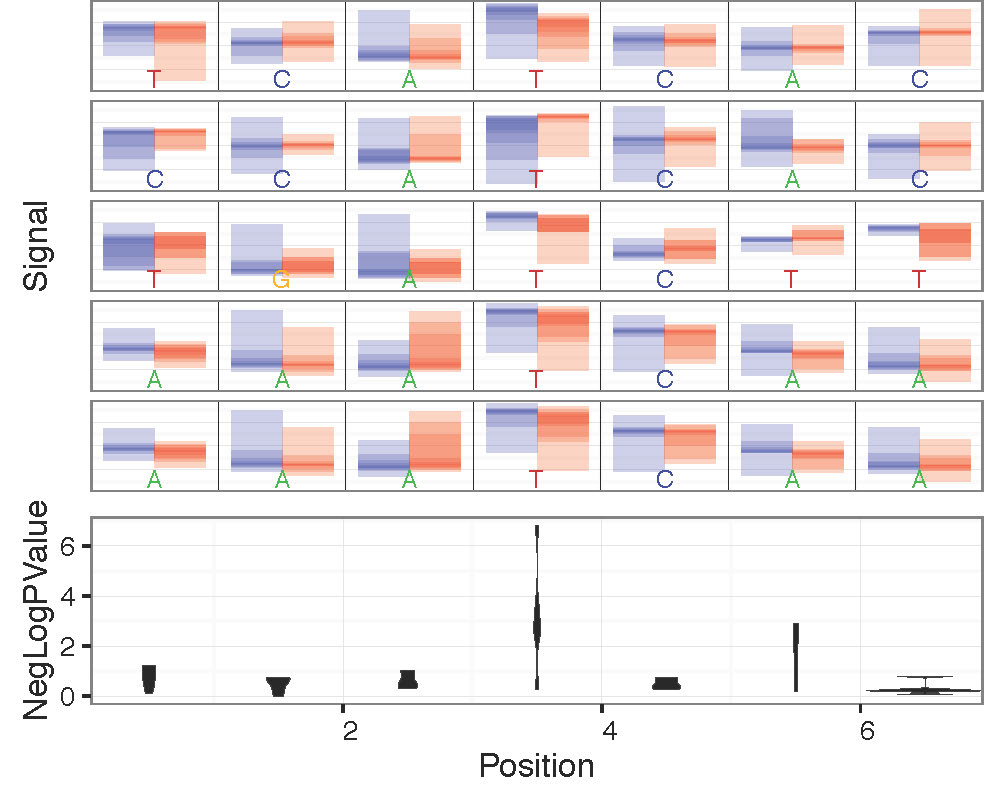
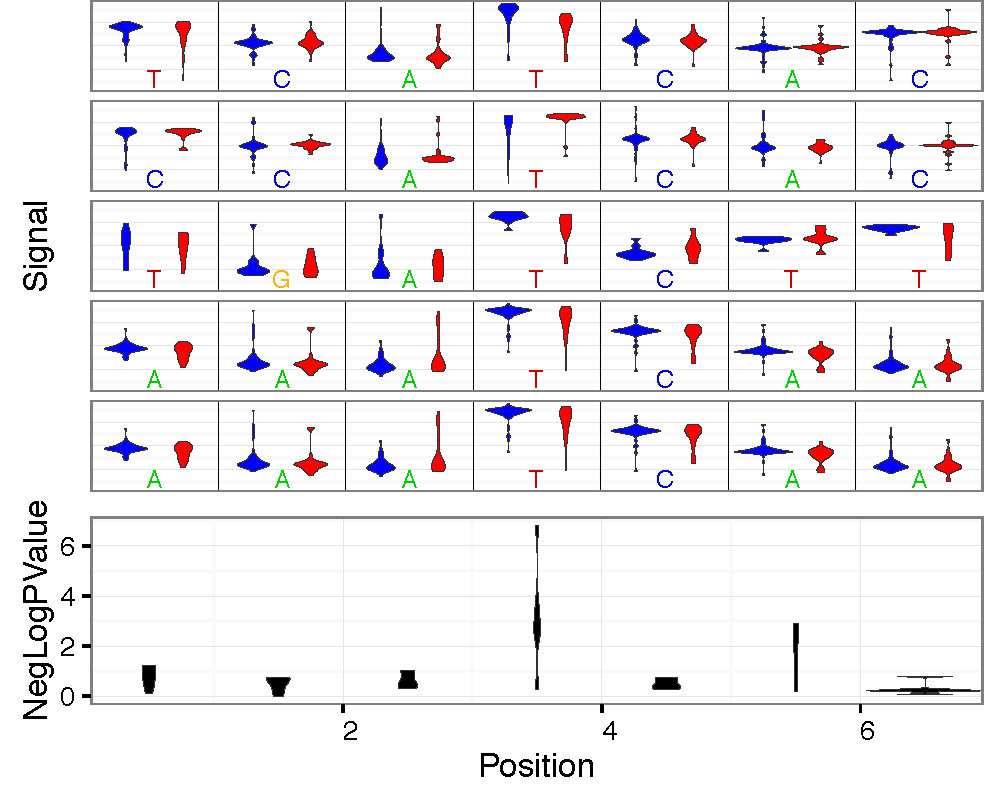
- –overplot-type: Plot type for regions with higher coverage. Choices: Downsample (default), Boxplot , Quantile, Violin. Examples below.
Data Slot Options¶
- –corrected-group: FAST5 group to access/plot created by genome_resquiggle script. Default: RawGenomeCorrected_000. The default is the default slot used by the genome_resquiggle command so this command will not need to be set unless you would like to access an alternatively re-squiggled slot (e.g. including multiple signal normalizations within the same file).
- –basecall-subgroups: FAST5 subgroup (under Analyses/[corrected-group]) where individual template and/or complement reads are stored. Default: BaseCalled_template. This is the default supplied by ONT and should work for most cases.
- –2d: Input contains 2D reads and both forward and complement should be plotted. Equivalent to –basecall-subgroups BaseCalled_template BaseCalled_complement
Command Specific Options¶
plot_genome_location Option¶
- –genome-locations: Plot signal at specified genomic locations. Regions will be centered on the specified genomic position. Format locations as “chrm:position [chrm2:position2 …]”. E.g. “chr1:1000 chr21:40000 chrY:5000”
plot_motif_centered and plot_motif_with_stats Options¶
- –motif: DNA motif of interest. Can be composed of any one letter DNA codes (NEB Single Letter Codes).
- –genome-fasta: FASTA file used to map reads with genome_resquiggle command. If chromosomes are missing then regions from those chromosomes (or organims if multi-species) will not be considered for plotting.
plot_most_significant and plot_motif_with_stats Options¶
- –test-type: Type of significance test to apply. Choices are: mw_utest (default; mann-whitney u-test), ttest.
- –fishers-method-offset: Offset up and downstream over which to compute combined p-values using Fisher’s method. For example 2 would compute the Fisher’s method p-value over a moving window of 5 bases. Default: Do not compute Fihser’s method p-values (report raw, base-by-base p-values).
- –statistics-filename: Filename to save/load base by base signal difference statistics. If file exists it will be loaded, if it does not exist it will be created to save statistics. Default: Don’t save/load. Note that –test-type and –fishers-method-offset will be ignored if –statistics-filename is provided and the file exists.
- –minimum-test-reads: Number of reads required from both samples to test for significant difference in signal level. Note that regions with lower coverage levels will not have p-values be computed. Default: 5
plot_most_significant Options¶
- –q-value-threshold: Choose the number of regions to plot by the FDR corrected p-values. Note that –num-regions will be ignored if this option is set.
- –sequences-filename: Filename to store genomic sequences at selected regions (e.g. for PWM search). Sequences will be stored in FASTA format. Default: None.
plot_motif_with_stats Option¶
- –num-context: Number of bases to plot surrounding motif of interest. Default: 2
Example commands¶
Single sample genome-anchored plotting functions:
nanoraw plot_max_coverage --fast5-basedirs $g1Dir --2d \
--num-bases 21 --overplot-threshold 1000
nanoraw plot_max_coverage --fast5-basedirs $g1Dir --2d \
--num-bases 21 --overplot-threshold 1000 \
--obs-per-base-filter 99:200 100:5000
nanoraw plot_genome_location --fast5-basedirs $g1Dir \
--genome-locations "S_aureus:2064835" "S_aureus:2064935" \
--2d --num-bases 21 --overplot-threshold 1000
nanoraw plot_motif_centered --fast5-basedirs $g1Dir --motif AHC \
--genome-fasta $genomeFn --2d \
--num-bases 21 --overplot-threshold 1000
nanoraw plot_motif_centered --fast5-basedirs $g1Dir --motif AHC \
--genome-fasta $genomeFn --2d \
--num-bases 21 --overplot-threshold 1000 --deepest-coverage
Mutliple sample genome-anchored plotting functions:
nanoraw plot_max_coverage --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir --2d \
--num-bases 21 --overplot-threshold 1000
nanoraw plot_max_coverage --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir --2d \
--num-bases 21 --overplot-threshold 1000 \
--obs-per-base-filter 99:200 100:5000
nanoraw plot_genome_location --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir \
--genome-locations "S_aureus:2064835" "S_aureus:2064935" \
--2d --num-bases 21 --overplot-threshold 1000
nanoraw plot_motif_centered --fast5-basedirs $g1Dir --motif AHC \
--genome-fasta $genomeFn \
--fast5-basedirs2 $g2Dir --2d \
--num-bases 21 --overplot-threshold 1000 --deepest-coverage
Mutliple sample statistical testing genome-anchored plotting functions:
nanoraw plot_max_difference --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir --2d \
--num-bases 21 --overplot-threshold 1000
nanoraw plot_most_significant --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir --2d \
--num-bases 21 --overplot-threshold 1000
nanoraw plot_motif_with_stats --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir --motif AHC --2d \
--overplot-threshold 1000 --test-type mw_utest \
--genome-fasta $genomeFn
Overplotting options:
nanoraw plot_max_coverage --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir --2d \
--num-bases 21 --overplot-threshold 20 --overplot-type Downsample \
--pdf-filename Nanopore_read_coverage.max_coverage.Downsample.pdf
nanoraw plot_max_coverage --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir --2d \
--num-bases 21 --overplot-threshold 20 --overplot-type Boxplot \
--pdf-filename Nanopore_read_coverage.max_coverage.Boxplot.pdf
nanoraw plot_max_coverage --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir --2d \
--num-bases 21 --overplot-threshold 20 --overplot-type Quantile \
--pdf-filename Nanopore_read_coverage.max_coverage.Quantile.pdf
nanoraw plot_max_coverage --fast5-basedirs $g1Dir \
--fast5-basedirs2 $g2Dir --2d \
--num-bases 21 --overplot-threshold 20 --overplot-type Violin \
--pdf-filename Nanopore_read_coverage.max_coverage.Violin.pdf